Researchers from the Jackson Laboratory contribute new scRNA-seq data on microglia in genetically diverse mouse models
By Zoë Leanza
“Imagine your brain as a fruit smoothie.”
Stanley Yang, of The Jackson Laboratory, was trying to explain why he and his co-investigators used single-cell RNA sequencing technology – instead of bulk – for their study of microglia of wild-derived mice. Data from this study was contributed as part of the AD Knowledge Portal Community Data Contribution Program, and can be found in the portal through the scRNAseq_microglia_wild_
“Bulk RNA sequencing is like taking a sip of the smoothie with all the flavors blended,” Yang said. “It provides information about a sample, but it cannot differentiate among cell types.”
Fellow Jackson Laboratory investigator Kristen Onos carried the analogy further: “Maybe the smoothie doesn’t taste right,” Onos said. “But you have no idea why, because everything is mixed together.”
Yang and Onos, with co-investigator Gareth Howell, provided single-cell RNA-sequencing data through the AD Knowledge Portal’s new Community Data Contribution Program. Their work supports efforts within the scientific community to tackle the complexity of a wide range of potential risk factors for Alzheimer’s disease (AD).
“Those risk factors might be immunological in some, and metabolic in others – or it could be something else entirely,” said Onos.
To mirror this diversity, the team generated data from mice with diverse genetic backgrounds. This was an important step that allowed them to examine how different genes might change the combination of cells in our brains or the way they behave. A change in brain cell composition could contribute to AD.
One important kind of brain cell is microglia, which act as the brain’s immune defense. Microglia clear out the sticky plaques that characterize AD, and they might be the “bad ingredient” in the smoothie analogy.
As described in their recent publication in Cell Reports, Yang and Onos found that the ability of some microglia to clear plaques normally could be impaired, as suggested by higher levels of AD associated genes like Apoe and Trem2.
By contributing raw single cell RNA sequencing data, processed data, and sample information to the AD Knowledge Portal, Yang and Onos have provided a reusable resource for the research community of these genetically diverse mouse strains. Qualified researchers can examine the raw data, perform downstream analysis, and retrieve information about microglia genes of interest using a user-friendly website.
Like most data available on the Portal, the data has been contributed for community use, encouraging early stage research without competing for intellectual property.
“I’m a huge advocate for open science,” Onos said. “Share everything as much as you can, because someone else might see something in your dataset that you didn’t even know to look for. And that could be the key to unlocking a whole new set of discoveries for this really complex disease.”
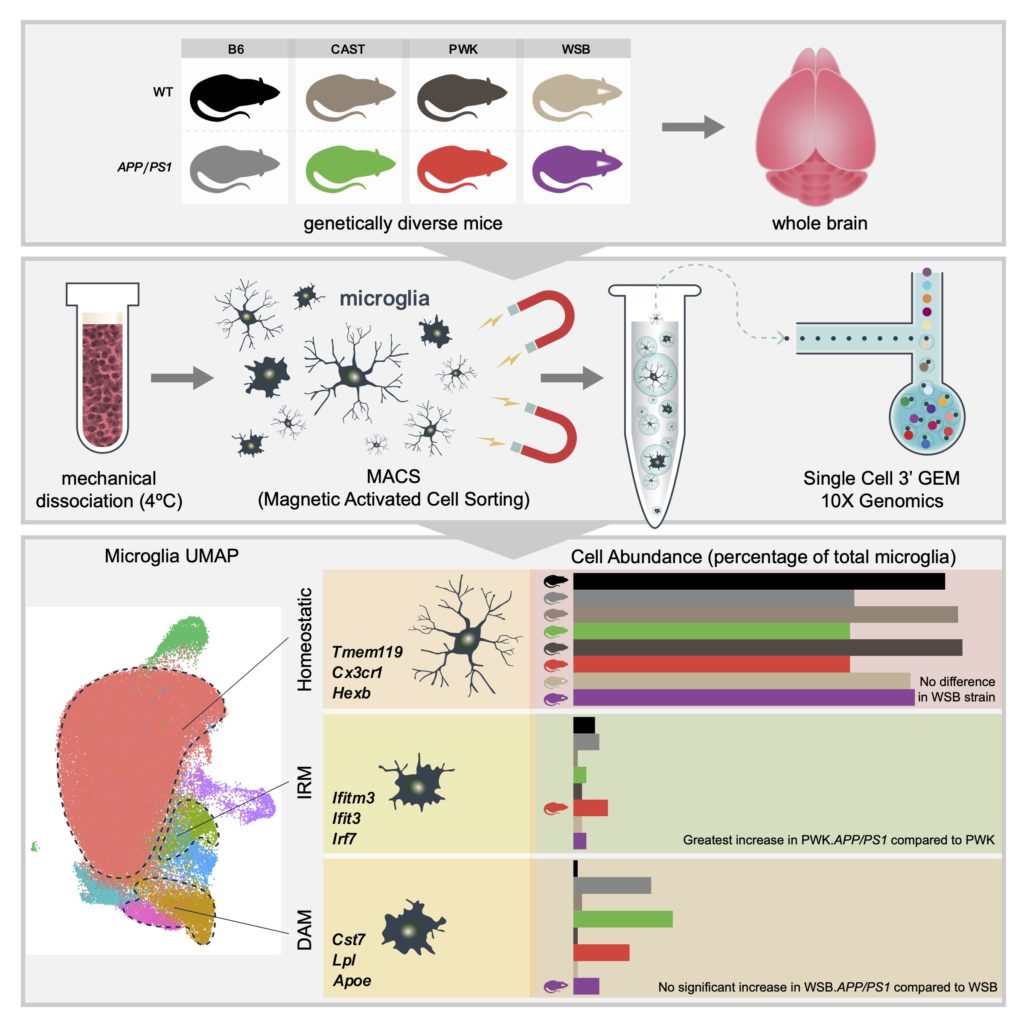
Difference in Microglia Abundance
Mice from four different genetic backgrounds with and without Alzheimer’s disease (AD) pathology were aged to 8 months and brains were collected for sequencing. Individual cells, microglia, were isolated from each brain, identified with a special tag and then sequenced together. All of the individual cells were clustered together based on similarity to identify a number of states of microglia: most were in a resting state (homeostatic), while some were interferon responding microglia (IRM), and others were disease associated microglia (DAM). It was discovered that genetic background and AD pathology led to differences in the abundance and gene expression of microglia.
More information
For access to content described in this manuscript see:
https://doi.org/10.7303/syn23763409.
For access to analytical code, see:
https://github.com/TheJacksonLaboratory/wild_AD_mic_scRNA.
More About CDCP
CDCP contributions are evaluated based on relevance to AD, complementarity to existing data, and unmet needs. If you have data, analyses, or tools that may benefit Alzheimer’s disease research, apply to become a community contributor.