Lab of Nilüfer Ertekin-Taner Discovers Glial DDR2, STOM, and KANK2 as Therapeutic Targets in Progressive Supranuclear Palsy
🧠 by Zoë Leanza
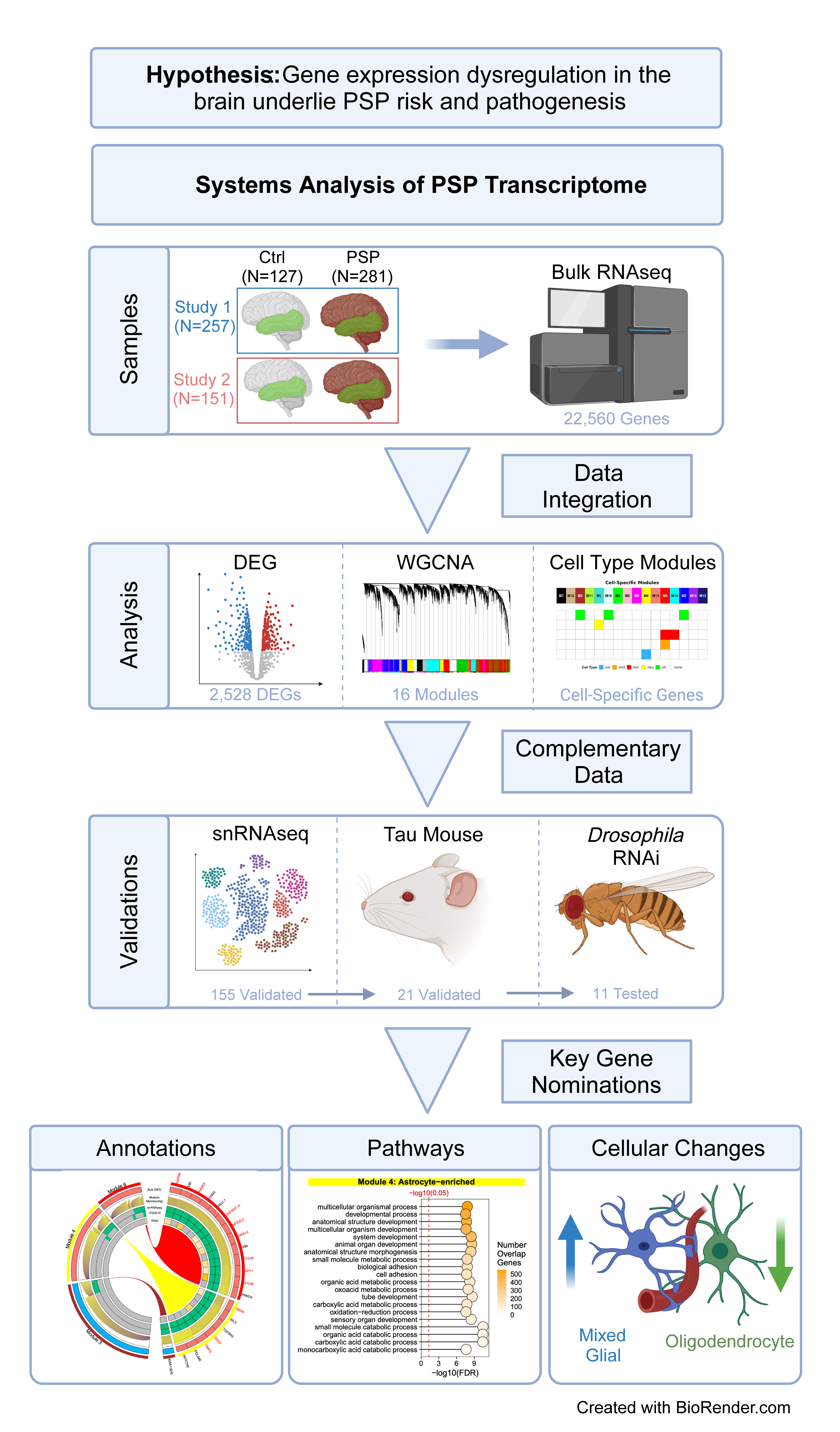
Three new therapeutic targets for progressive supranuclear palsy (PSP) have been identified by the lab of Nilüfer Ertekin-Taner, M.D., Ph.D., FAAN at Mayo Clinic Florida. Affecting just six people per 100,000, PSP is an uncommon brain disorder that can disrupt body movements and balance.
Recognizing a major gap in data availability, the team at Dr. Nilüfer Ertekin-Taner’s lab also contributed the data behind their own findings to the AD Knowledge Portal. By sharing this extensive transcriptomic dataset, the researchers are accelerating the work of others in the field, empowering the scientific community to pursue solutions faster.

The Lab of Dr. Nilüfer Ertekin-Taner
“PSP is an incurable, rapidly progressing neurodegenerative disorder characterized by cell-type specific tau lesions,” says Yuhao (Harry) Min, a graduate student in Ertekin-Taner’s lab. “Our work uses a multi-omics approach to understand the complex biology behind it.”
The researchers aim to uncover the molecular changes that occur during the development of PSP, pinpoint the affected brain cell types, and identify potential targets for therapy. They analyzed brain samples from over 400 individuals, generating bulk tissue RNAseq and single-nucleus RNAseq data from both patients with PSP and healthy controls.
In PSP-affected brains, the team observed significant changes in gene expression patterns, including the down-regulation of myelin-related genes and the up-regulation of cell metabolism and immune-related genes.
With thousands of genes affected in PSP, prioritizing the targets was no easy feat. The team developed a multi-stage, cross-species selection process to maximize the likelihood of discovering potential candidates.
They used RNAseq data from humans and mouse models in this process to produce a short list of 11 genes that are implicated in PSP. Next, using an experimental approach with a fruit fly model of tauopathy, the team identified three of the most promising candidate genes with the strongest rescue of tau-mediated toxicity – DDR2, KANK2, and STOM, which may indicate downstream therapeutic significance for PSP. By focusing on these unique targets, this research could lead to more effective treatments for patients with PSP.
The contributed dataset, one of the largest of its kind, not only serves as a valuable resource for advancing PSP research, but also holds promise for research in other fields. Researchers can access and download the data to address questions about other neurodegenerative disorders, such as Alzheimer’s disease. For example, because PSP is primarily characterized by tauopathy, researchers can examine commonalities with AD, which has both tau and amyloid pathologies.
To make their findings even more accessible, the team developed a user-friendly web application. This resource allows researchers to explore the data conveniently, even from a mobile device. “So the next time you are at a seminar, and an interesting gene pops up,” Min says, “you can quickly look it up on your phone and see how it changes in PSP.”
As Min points out, there is an incredible amount of biologically relevant information in multi-omics data. “I am amazed that we can profile the expression levels of tens of thousands of genes simultaneously, sometimes even at a single-cell resolution,” he says, “This is so powerful as it captures the changes in biological systems and allows us to really understand what is going on.”
In addition to the team’s contribution of pathologically confirmed PSP RNAseq data, the introduction of a novel pipeline may help the research community leverage data and tools for therapeutic development. The direct nomination of top genes provides direction for the scientific community, illuminating the need for research into relevant pathways and genes. Finally, the use-friendly web application enables quick research into genes of interest, bringing the community closer to potential treatments for PSP and offering hope for patients.
More information
More information is available from the Mayo Clinic on the Genetics of Alzheimer’s Disease and Endophenotypes Laboratory and Dr. Nilüfer Ertekin-Taner.